* rewrite mocksnakemake for parsing real Snakefile * continue add function to scripts * going through all scripts, setting new mocksnakemake * fix plotting scripts * fix build_country_flh * fix build_country_flh II * adjust config files * fix make_summary for tutorial network * create dir also for output * incorporate suggestions * consistent import of mocksnakemake * consistent import of mocksnakemake II * Update scripts/_helpers.py Co-Authored-By: euronion <42553970+euronion@users.noreply.github.com> * Update scripts/_helpers.py Co-Authored-By: euronion <42553970+euronion@users.noreply.github.com> * Update scripts/_helpers.py Co-Authored-By: euronion <42553970+euronion@users.noreply.github.com> * Update scripts/_helpers.py Co-Authored-By: euronion <42553970+euronion@users.noreply.github.com> * Update scripts/plot_network.py Co-Authored-By: euronion <42553970+euronion@users.noreply.github.com> * Update scripts/plot_network.py Co-Authored-By: euronion <42553970+euronion@users.noreply.github.com> * Update scripts/retrieve_databundle.py Co-Authored-By: euronion <42553970+euronion@users.noreply.github.com> * use pathlib for mocksnakemake * rename mocksnakemake into mock_snakemake * revert change in data * Update scripts/_helpers.py Co-Authored-By: euronion <42553970+euronion@users.noreply.github.com> * remove setting logfile in mock_snakemake, use Path in configure_logging * fix fallback path and base_dir fix return type of make_io_accessable * reformulate mock_snakemake * incorporate suggestion, fix typos * mock_snakemake: apply absolute paths again, add assertion error *.py: make hard coded io path accessable for mock_snakemake * retrieve_natura_raster: use snakemake.output for fn_out * include suggestion * Apply suggestions from code review Co-Authored-By: Jonas Hörsch <jonas.hoersch@posteo.de> * linting, add return ad end of file * Update scripts/plot_p_nom_max.py Co-Authored-By: Jonas Hörsch <jonas.hoersch@posteo.de> * Update scripts/plot_p_nom_max.py fixes #112 Co-Authored-By: Jonas Hörsch <jonas.hoersch@posteo.de> * plot_p_nom_max: small correction * config.tutorial.yaml fix snapshots end * use techs instead of technology * revert try out from previous commit, complete replacing * change clusters -> clusts in plot_p_nom_max due to wildcard constraints of clusters * change clusters -> clusts in plot_p_nom_max due to wildcard constraints of clusters II |
||
|---|---|---|
| data | ||
| doc | ||
| scripts | ||
| test | ||
| .gitattributes | ||
| .gitignore | ||
| .readthedocs.yml | ||
| .travis.yml | ||
| borg-it | ||
| cluster.yaml | ||
| config.default.yaml | ||
| config.tutorial.yaml | ||
| environment.docs.yaml | ||
| environment.fixedversions.yaml | ||
| environment.yaml | ||
| LICENSE.txt | ||
| matplotlibrc | ||
| README.md | ||
| Snakefile | ||
PyPSA-Eur: An Open Optimisation Model of the European Transmission System
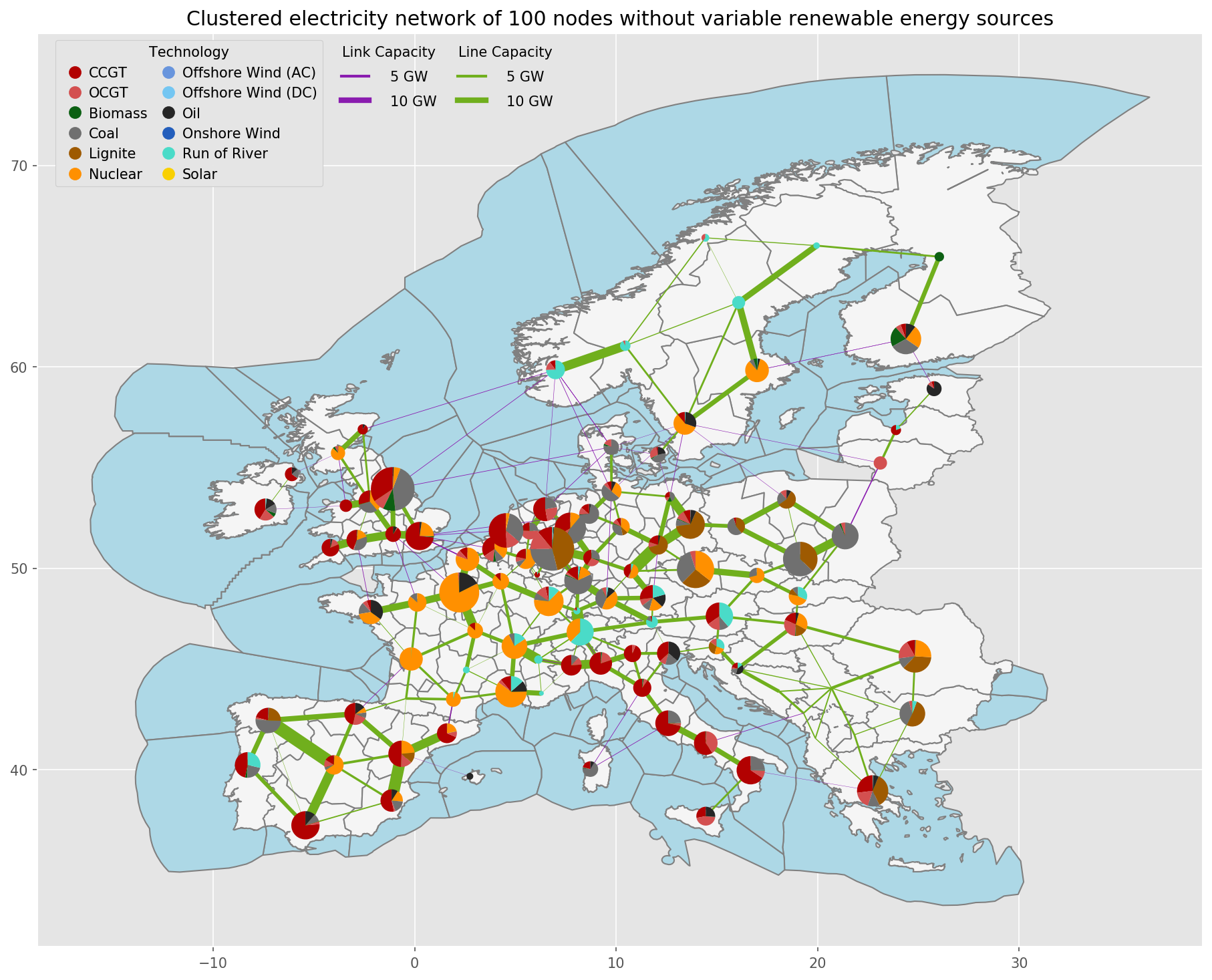
PyPSA-Eur is an open model dataset of the European power system at the transmission network level that covers the full ENTSO-E area. The model is suitable both for operational studies and generation and transmission expansion planning studies. The continental scope and highly resolved spatial scale enables a proper description of the long-range smoothing effects for renewable power generation and their varying resource availability.
The model is described in the documentation and in the paper PyPSA-Eur: An Open Optimisation Model of the European Transmission System, 2018, arXiv:1806.01613.
The model is designed to be imported into the open toolbox PyPSA for operational studies as well as generation and transmission expansion planning studies.
The dataset consists of:
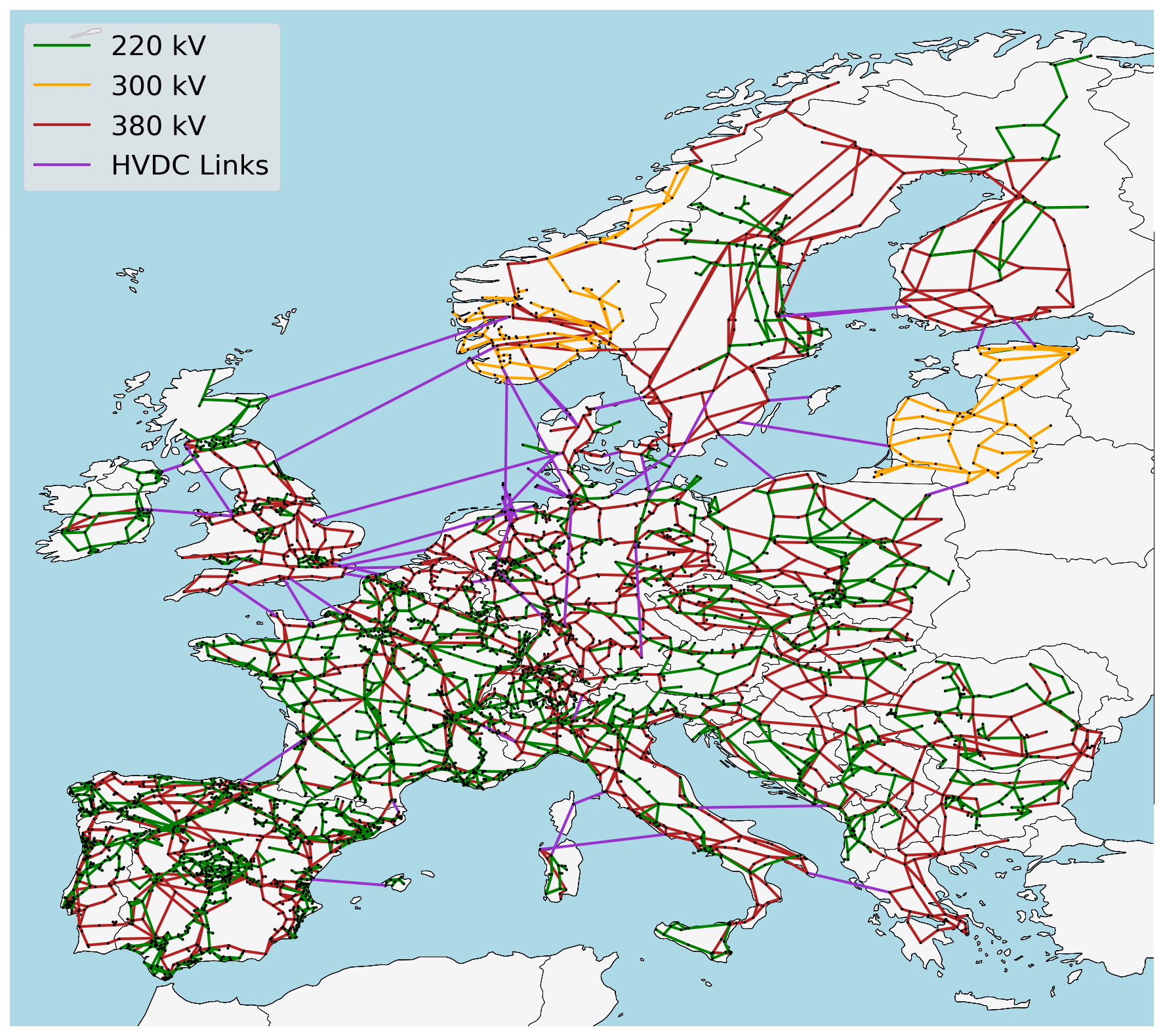
- A grid model based on a modified GridKit extraction of the ENTSO-E Transmission System Map. The grid model contains 6001 lines (alternating current lines at and above 220kV voltage level and all high voltage direct current lines) and 3657 substations.
- The open power plant database powerplantmatching.
- Electrical demand time series from the OPSD project.
- Renewable time series based on ERA5 and SARAH, assembled using the atlite tool.
- Geographical potentials for wind and solar generators based on land use (CORINE) and excluding nature reserves (Natura2000) are computed with the vresutils library and the glaes library.
Already-built versions of the model can be found in the accompanying Zenodo repository.